Aims and objectives
MRI control in patients after surgical treatment of glioblastoma is an important neuroimaging method,
both for assessing the volume of performed surgery and for detecting residual tumor tissue.
Glioblastoma growth is often accompanied by impaired blood-brain barrier due to its infiltrative growth and is characterized by elevated hemodynamic parameters (CBV,
CBF,
MTT),
associated with pronounced tumor angiogenesis (1,2,3).
However,
in the area of surgical intervention,
the destruction of the blood-brain barrier is also noted.
In this way the accumulation of a contrast agent in both...
Methods and materials
56 patients were included — 32 men (57.1%) and 24 women (42.9%) after 2-4 weeks surgical removal of glioblastoma (WHO GRADE IV),
prior to the course of radiotherapy.
The average age of patients was 56.9 ± 10.9 years (min- 31 years,
max- 79 years),
there were no significant differences in age between the examined men and women (56.1 ± 11.2 years in men,
58.1 ± 10.8 years in women,
p = 0.45).
MRI studies were performed on a 1.5T magnetic induction device (Optima MR450w,
GE...
Results
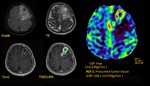
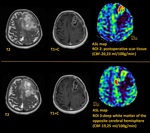
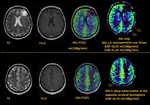
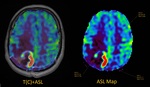
As a result of the study all patients were divided into two groups depending on the CBF value.
1st group - 38 patients (67.9%) with a pathological increase in cerebral blood flow on ASL perfusion cards (presumably a tumor – ROI 1),
the average CBF was 137.6 ± 35.2 ml/100g/min (minimum - 79.6 ml/100g/min,
max - 227.6 ml/100g/min) (Fig.2).
In this patients the CBF value of the supposed tumor site was 5-6 times higher than the blood flow in the area of postoperative scars (ROI...
Conclusion
In our study we showed the CBF value in tumor tissue is significantly higher than CBF in postoperative scar tissue.
In this way the capacity of ASL perfusion is sufficient to identify residual tumor tissue in patients after surgical treatment of high-grade gliomas that have an aggressive course and expressed angiogenesis,
it can be a crucial step in choosing an approach to further treatment and radiotherapy planning.
Personal information
M.
Bunak,
Dr.
Moscow Regional Research and Clinical Institute ("MONIKI"),
Department of Radiology
Schepkina str.
61/2 k.1
PO Box 129110
Moscow,
Russian Federation
Phone: +79264374191
E-mail :
[email protected]
References
1.
Leon SP,
Folkerth RD,
Black PM: Microvessel density is a prognostic indicator for patients with astroglial brain tumors.
Cancer 77: 362 372,
1996.
2.
Huang AP,
Tsai JC,
Kuo LT,
Lee CW,
Lai HS,
Tsai LK,
Huang SJ,
Chen CM,
Chen YS,
Chuang HY,
Wintermark M: Clinical application of perfusion computed tomography in neurosurgery.
J Neurosurg 120: 473 488,
2014.
3.
Jain R.
Perfusion CT imaging of brain tumors: an overview.
American Journal of Neuroradiology.
October 2011;32(9):1570–1577
4.
Knauth,
M.,
Aras,
N.,
Wirtz,
C.R.,...