Aims and objectives
This study presents a platform for the longitudinal analysis of the DW-MRI data.
A graphical user interface (GUI) empowers users to read,
analyze,
visualize and quantify the data into diffusion related parameters using models with variable degree of complexity (Gaussian and non-Gaussian mono- and bi-exponential models).
The platform follows a pipeline workflow comprising: a) data import and preparation for further analysis,
b) quantification of diffusion related imaging biomarkers based on different models,
c) qualitative assessment by visual inspection of colour coded parametric maps,
d) statistical...
Methods and materials
Import raw medical imaging data
The platform supports image import from most widely used MRI scanners,
and accesses the DICOM header in order to retrieve information on the acquisition protocol.
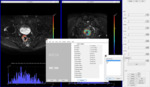
Region of Interest (ROI) delineation
Once data are imported properly,
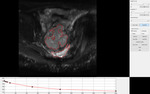
the platform allows interaction with the user for selecting the ROI(s) for analysis Fig. 1.
ROI delineation is performed either manually by contour drawing within the slices or by importing the ROI(s) that are stored locally as medical imaging files (i.e.
NIFTI format).
Manual drawing...
Results
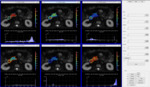
A single or longitudinal analysis can be displayed using parametric maps for all model related parameters (ADC,
f,
D,
D*,
fxD,
fxD*,
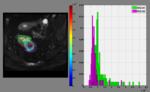
and K) in a pixel-by-pixel basis (Fig. 2).
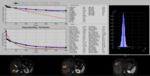
All studies are coupled with evaluation criteria based on b-value specific SNR maps.The minimum and maximum value of the desired metric can also be adjusted by the user,
as well as the tolerance of the fitting algorithm in order to deactivate visualization of pixels with poor fitting performance.
The delineation of a smaller ROI inside...
Conclusion
A comprehensive software tool for analyzing DW-MRI data is presented.
It is structured in a simple workflow allowing minimal interaction with non-experienced users but at the same time it represents a complete research tool in the hands of more experienced users in medical analysis and visualization.
In both cases it succeeds maximum knowledge extraction from a vast pool of information,
in a simple and robust display scheme.
Personal information
Name: G.C.
Manikis,
Electronic & Computer Engineer,
MSc
Department: Computational BioMedicine Laboratory (CBML),
Institute of Computer Science (ICS),
Foundation for Research and Technology - Hellas (FORTH)
Email:
[email protected]
References
J.H.
Burdette,
A.D.
Elster,
P.E.
Ricci.
1998.
Calculation of apparent diffusion coefficients (ADCs) in brain using two-point and six-point methods.
J Comput Assist Tomogr; 22:792–794.
D.
Le Bihan,
E.
Breton,
D.
Lallemand,
M.-L.
Aubin,
J.
Vignaud,
and M.
Laval-Jeantet.
1988.
Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging.
Radiology,
vol.
168,
no.
2,
pp.
497–505.
A.
L.
Sukstanskii,
D.
A.
Yablonskiy,
and J.
J.
H.
Ackerman.
2004.
Effects of permeable boundaries on the diffusion-attenuated MR signal: insights from a one-dimensional model.
J....