Keywords:
Oedema, Computer Applications-General, Computer Applications-Detection, diagnosis, Digital radiography, Cardiovascular system, Artificial Intelligence
Authors:
J. W. Luo, J. J. R. Chong; Montreal, QC/CA
DOI:
10.26044/ecr2019/C-3644
Methods and materials
A retrospective case-control review on McGill PACS isolated 1346 chest radiographs consisting of 673 cases positive for cardiomegaly and 673 controls.
Cases and controls were manually selected,
balanced and filtered to keep studies with posteroanterior (PA) views only.
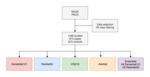
Four different convolutional neural network architectures,
AlexNet,
VGG19,
ResNet50,
and DenseNet121,
were compared.
The two newest architectures were further ensembled in order to measure the impact of model ensembling on final performance (Figure 1).
The balanced dataset was split 80% training / 20% validation,
corresponding to 1120 training and 226 validation cases respectively. Images underwent histogram normalization and standard affine data augmentation techniques (random cropping,
rotation,
shearing,
and horizontal flipping).
Each network was trained for 200 epochs using stochastic gradient descent (SGD) with a starting learning rate of 0.001.
A learning rate decay factor of 10 was applied after 100 epochs.
Network training was done with and without transfer learning from pre-trained weights on the 112,120 deidentified chest radiographs from the NIH dataset [2].
Areas under the receiver operating characteristic (AUC) curves assessed model performance.
Reported AUC values were those of the training runs with the lowest cross-entropy validation loss on each neural network.