Institutional review board approval was obtained from Marmara University Hospital,
Istanbul,
Turkey and all subjects signed the informed consent before data acquisition.
MRI images of 69 patients were obtained in prone and supine positions.
Standard clinical breast MRI images were acquired using a 3-Tesla scanner with the patient in prone position with a 16-channel phased-array dedicated breast coil.
Examination protocol included following sequences: (1) Axial turbo spin-echo fat saturated T2-weighted sequence (TR/TE 4100/70 ms,
field of view 30 cm,
acquisition matrix 440x380,
slice thickness 3 mm,
and acquisition time 3 minutes 43 seconds).
(2) Diffusion-weighted imaging using echo-planar image (EPI) sequence with fat suppression (b values 50,
400 and 800 s/mm2,
TR/TE 9700/86 ms,
FOV 30 cm,
in-plane resolution 1.7x2 mm2,
slice thickness 3 mm,
and acquisition time 4 minutes 32 seconds).
(3) Three-dimensional (3D) volumetric interpolated (VIBE) sequence (TR/TE 5.01/1.77 ms,
FOV 30 cm,
acquisition matrix 512x460,
slice thickness 1 mm,
in-plane resolution 0.6x0.7x1.0 mm,
and total acquisition time 10 minutes with a temporal resolution of 82 s) for dynamic contrast enhanced sequence.
The Gadolinium based contrast agent was administered intravenously via an antecubital vein at a rate of 2 mL/s using an automated injector system followed by a saline injection.
Subtracted contrast-enhanced dynamic images were used as standard for lesion identification.
After completion of the prone breast MR imaging,
the patient was taken out of the magnet.
The dedicated breast coil was retrieved.
Patient position was changed to supine with both arms extended over the head.
A flexible 6 channel body coil was centralized on the sternum and placed over both breasts and was fixed with a strap to prevent coil displacement and minimize artifacts due to respiratory motion.
A single 3D turbo field echo sequence was used for T1-weighted high-resolution isotropic examination volume without fat suppression (TR/TE = 2030/12 ms; flip angle = 120 degrees; field of view = 320 mm; matrix = 322 × 480 mm; acquisition time = 82sec).
The prone and supine images in each set were registered using common reference points.
Two trained radiologists,
with more than 10 years of experience in breast imaging,
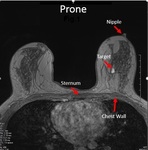
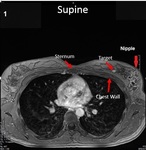
hand-selected and recorded the position of the target lesions and eight landmarks in prone and supine positions,
at the chest wall and anterior breast surface including the nipple and other skin points.
(Figures 1 and 2).
Additional predictors included breast volume and breast composition on a scale of 1 to 4,
from the least to most fibro-glandular tissue to fatty tissue ratio.
From this MRI analysis,
a dataset of positional coordinates and other predictors was created from 101 observations,
each corresponding to one target and the associated predictors.
The position coordinates of targets and landmarks,
as well as derivate measurements representing distances,
distance ratios,
and vector angles were calculated.
Of the 69 patients,
25 had multiple targets and 44 had single targets,
which ranged in size between 5 to 25 mm in largest diameter.
Our ML predictive model is based on the Elastic Net algorithm [3] ,
a linear regression model with both L1 and L2 penalties; these penalties result in a built-in feature selection that works well on data sets with many predictors/few observations as well as highly correlated predictors.
The dataset was trained on this algorithm using Scikit-Learn Python [4],
and the prediction accuracy was assessed using 4-fold cross-validation with data only from subjects with single targets used for accuracy assessment to avoid data leakage.